RamiGO 安装及库依赖解决备忘
一、RamiGO 安装¶
$ /home/shenweiyan/software/R/R-3.6.1/bin/R CMD INSTALL /Bioinfo/Pipeline/SourceCode/pkgs/RamiGO_1.20.0.tar.gz
* installing to library ‘/home/shenweiyan/software/R/R-3.6.1/lib64/R/library’
ERROR: dependencies ‘gsubfn’, ‘igraph’, ‘RCurl’, ‘png’, ‘RCytoscape’, ‘graph’ are not available for package ‘RamiGO’
* removing ‘/home/shenweiyan/software/R/R-3.6.1/lib64/R/library/RamiGO’
# 安装 gsubfn, igraph, RCurl
install.packages(c("igraph", "igraph", "RCurl", "png"))
# 安装 graph
BiocManager::install("graph")
$ /home/shenweiyan/software/R/R-3.6.1/bin/R CMD INSTALL /Bioinfo/Pipeline/SourceCode/pkgs/RCytoscape_1.12.0.tar.gz
* installing to library ‘/home/shenweiyan/software/R/R-3.6.1/lib64/R/library’
ERROR: dependency ‘XMLRPC’ is not available for package ‘RCytoscape’
* removing ‘/home/shenweiyan/software/R/R-3.6.1/lib64/R/library/RCytoscape’
install.packages("devtools")
install_github("duncantl/XMLRPC")
$ /home/shenweiyan/software/R/R-3.6.1/bin/R CMD INSTALL /Bioinfo/Pipeline/SourceCode/pkgs/RamiGO_1.20.0.tar.gz
* installing to library ‘/home/shenweiyan/software/R/R-3.6.1/lib64/R/library’
* installing *source* package ‘RamiGO’ ...
** using staged installation
** R
** data
** inst
** byte-compile and prepare package for lazy loading
Error in dyn.load(file, DLLpath = DLLpath, ...) :
unable to load shared object '/home/shenweiyan/software/R/R-3.6.1/lib64/R/library/png/libs/png.so':
libpng16.so.16: cannot open shared object file: No such file or directory
Calls: <Anonymous> ... asNamespace -> loadNamespace -> library.dynam -> dyn.load
Execution halted
ERROR: lazy loading failed for package ‘RamiGO’
* removing ‘/home/shenweiyan/software/R/R-3.6.1/lib64/R/library/RamiGO’
$ export LD_LIBRARY_PATH=/home/shenweiyan/software/LibDependence/libpng-1.6.37/
bin/ include/ lib/ share/
$ export LD_LIBRARY_PATH=/home/shenweiyan/software/LibDependence/libpng-1.6.37/lib:$LD_LIBRARY_PATH
$ /home/shenweiyan/software/R/R-3.6.1/bin/R CMD INSTALL /Bioinfo/Pipeline/SourceCode/pkgs/RamiGO_1.20.0.tar.gz
* installing to library ‘/home/shenweiyan/software/R/R-3.6.1/lib64/R/library’
* installing *source* package ‘RamiGO’ ...
** using staged installation
** R
** data
** inst
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
** building package indices
** installing vignettes
** testing if installed package can be loaded from temporary location
** testing if installed package can be loaded from final location
** testing if installed package keeps a record of temporary installation path
* DONE (RamiGO)
二、RamiGO 使用与 libpng 依赖库异常¶
使用 RamiGO R 包过程中,如果发现:
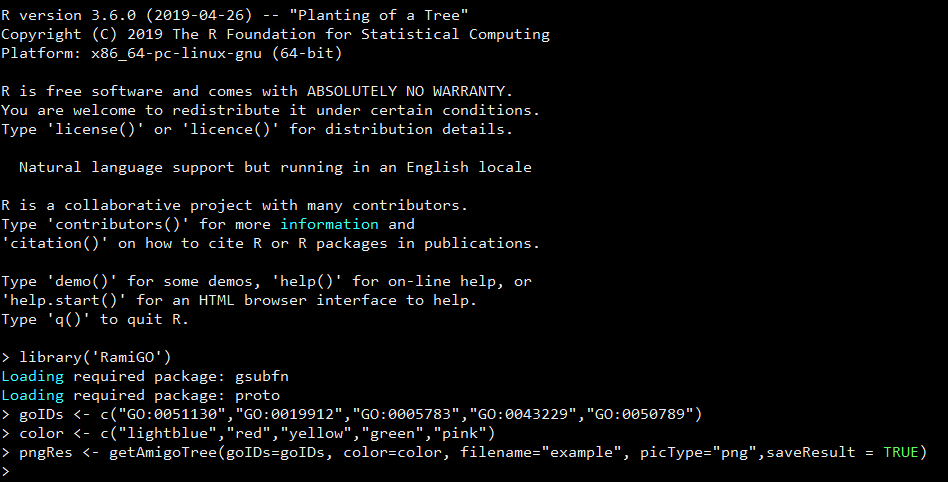
> library('RamiGO')
Loading required package: gsubfn
Loading required package: proto
> goIDs <- c("GO:0051130","GO:0019912","GO:0005783","GO:0043229","GO:0050789")
> color <- c("lightblue","red","yellow","green","pink")
> getAmigoTree(goIDs=goIDs, color=color, filename="example", picType="png",saveResult = TRUE)
Error in readPNG(aa) :
libpng error: Incompatible libpng version in application and library
In addition: Warning messages:
1: In readPNG(aa) :
libpng warning: Application was compiled with png.h from libpng-1.6.37
2: In readPNG(aa) :
libpng warning: Application is running with png.c from libpng-1.2.49
首先,如果是源码编译安装的 R(参考:《手把手教你如何在 Linux 源码安装最新版本的 R》),可以尝试在 configure 的时候把 libpng 相关的的 LDFLAGS 和 CPPFLAGS 去掉,重新安装 R。
然后,通过拷贝缺失的共享库解决存在的问题。
> library(RamiGO)
Loading required package: gsubfn
Loading required package: proto
Error: package or namespace load failed for ‘RamiGO’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/shenweiyan/software/R/R-3.6.1/lib64/R/library/png/libs/png.so':
libpng16.so.16: cannot open shared object file: No such file or directory
> goIDs <- c("GO:0051130","GO:0019912","GO:0005783","GO:0043229","GO:0050789")
> color <- c("lightblue","red","yellow","green","pink")
> getAmigoTree(goIDs=goIDs, color=color, filename="example", picType="png",saveResult = TRUE)
Error in getAmigoTree(goIDs = goIDs, color = color, filename = "example", :
could not find function "getAmigoTree"
$ ldd /home/shenweiyan/software/R/R-3.6.1/lib64/R/library/png/libs/png.so
linux-vdso.so.1 => (0x00007fff48999000)
libpng16.so.16 => not found
libm.so.6 => /lib64/libm.so.6 (0x00007f100a740000)
libz.so.1 => /RiboBio/Bioinfo/APPS/R-3.3.2/lib/libz.so.1 (0x00007f100a528000)
libR.so => not found
libc.so.6 => /lib64/libc.so.6 (0x00007f100a194000)
/lib64/ld-linux-x86-64.so.2 (0x0000003636a00000)
$ ln -s /home/shenweiyan/software/LibDependence/libpng-1.6.37/lib/libpng16.so.16.37.0 /usr/lib64/libpng16.so.16
$ ln -s /home/shenweiyan/software/R/R-3.6.1/lib64/R/lib/libR.so /usr/lib64/libR.so
$ ldd /home/shenweiyan/software/R/R-3.6.1/lib64/R/library/png/libs/png.so
linux-vdso.so.1 => (0x00007fff079ff000)
libpng16.so.16 (0x00007fd903bed000)
libm.so.6 => /lib64/libm.so.6 (0x00007fd903969000)
libz.so.1 => /Bioinfo/APPS/R-3.3.2/lib/libz.so.1 (0x00007fd903751000)
libR.so => /usr/lib64/libR.so (0x00007fd9030aa000)
libc.so.6 => /lib64/libc.so.6 (0x00007fd902d16000)
libRblas.so => not found
libgfortran.so.3 => /usr/lib64/libgfortran.so.3 (0x00007fd902a23000)
libreadline.so.6 => /lib64/libreadline.so.6 (0x00007fd9027e0000)
libpcre.so.1 => /Bioinfo/APPS/R-3.3.2/lib/libpcre.so.1 (0x00007fd9025ad000)
liblzma.so.5 => /Bioinfo/APPS/R-3.3.2/lib/liblzma.so.5 (0x00007fd902388000)
librt.so.1 => /lib64/librt.so.1 (0x00007fd902180000)
libdl.so.2 => /lib64/libdl.so.2 (0x00007fd901f7b000)
libiconv.so.2 => /usr/local/lib/libiconv.so.2 (0x00007fd901c96000)
libgomp.so.1 => /Bioinfo/APPS/gcc-5.1.0/lib64/libgomp.so.1 (0x00007fd901a77000)
libpthread.so.0 => /lib64/libpthread.so.0 (0x00007fd901859000)
/lib64/ld-linux-x86-64.so.2 (0x0000003636a00000)
libtinfo.so.5 => /lib64/libtinfo.so.5 (0x00007fd901638000)
$ ln -s /home/shenweiyan/software/R/R-3.6.1/lib64/R/lib/libRblas.so /usr/lib64/libRblas.so
$ ldd /home/shenweiyan/software/R/R-3.6.1/lib64/R/library/png/libs/png.so
linux-vdso.so.1 => (0x00007fff2b7ff000)
libpng16.so.16 (0x00007fed3cd93000)
libm.so.6 => /lib64/libm.so.6 (0x00007fed3cb0f000)
libz.so.1 => /Bioinfo/APPS/R-3.3.2/lib/libz.so.1 (0x00007fed3c8f7000)
libR.so => /usr/lib64/libR.so (0x00007fed3c250000)
libc.so.6 => /lib64/libc.so.6 (0x00007fed3bebc000)
libRblas.so => /usr/lib64/libRblas.so (0x00007fed3bc90000)
libgfortran.so.3 => /usr/lib64/libgfortran.so.3 (0x00007fed3b99e000)
libreadline.so.6 => /lib64/libreadline.so.6 (0x00007fed3b75b000)
libpcre.so.1 => /Bioinfo/APPS/R-3.3.2/lib/libpcre.so.1 (0x00007fed3b528000)
liblzma.so.5 => /Bioinfo/APPS/R-3.3.2/lib/liblzma.so.5 (0x00007fed3b303000)
librt.so.1 => /lib64/librt.so.1 (0x00007fed3b0fb000)
libdl.so.2 => /lib64/libdl.so.2 (0x00007fed3aef6000)
libiconv.so.2 => /usr/local/lib/libiconv.so.2 (0x00007fed3ac11000)
libgomp.so.1 => /Bioinfo/APPS/gcc-5.1.0/lib64/libgomp.so.1 (0x00007fed3a9f2000)
libpthread.so.0 => /lib64/libpthread.so.0 (0x00007fed3a7d4000)
/lib64/ld-linux-x86-64.so.2 (0x0000003636a00000)
libtinfo.so.5 => /lib64/libtinfo.so.5 (0x00007fed3a5b3000)